Upcoming Virtual dSTORM Training – Coming January 29, 2026 register now>
Understanding the brain at different scales: from tissue to single molecules
Neuroscience Hub
Single-molecule imaging for neuroscience
The human brain is one of the most complex systems in biology. With its 100 trillion connections, the neuronal network controls motor responses and sensory information that determine our memory, identity, consciousness and creativity. Understanding how the brain functions at a single-molecule level is key for understanding the foundations of neurodegenerative disorders, including Alzheimer’s and Parkinson’s disease. The mechanisms underlying
brain degeneration onset remain largely unresolved; current efforts are focusing on understanding its molecular hallmarks to improve early diagnosis and develop more efficient drugs. This guide summarizes how single-molecule imaging is unlocking the understanding of brain function in health and disease.
Chapter 1
Studying neuronal connections
Brain functions are operated through neural connections between dendritic spines and axon terminals, known as synapses. Synapses operate in a sophisticated and cooperative manner in order to maintain brain function and adapt/respond to stimuli.
Challenges in understanding synapses
The small size of synapses, under 200 nm in size, can hinder the study of the molecular mechanisms regulating neurotransmission. Each synapse is composed of over 200 proteins and a synaptic cleft, across which communication between cell terminals is mediated, that can be as small as 20-40nm (Figure 1).
Understanding the crucial mechanisms that determine neuronal connections in health and disease is limited by multiple factors. Firstly, culture cell lines for certain brain cell types can be difficult to obtain and maintain in vitro, making it challenging to find truly predictive neural models. Currently, the most accessible super-resolution imaging studies use primary cortical and hippocampal cell cultures from mice or rats. Yet these models can be considered artificial environments with low complexity brain architectures. An attractive alternative for neuroscientists is the use of neurons differentiated from iPSCs (induced Pluripotent Stem Cells, generated directly from adult cells), which allows research on human cell lines. However, the lack of disease models necessary for iPCS is limiting research using this approach.
Another limitation in neuroimaging is the availability of labeling tools; available antibodies target the most common and best researched proteins while other players remain unexplored, hindering the detailed analysis of neuronal synapse architecture. To learn more on neuronal culture preparation and labeling read our Technical guide on Neuronal Culture for Super-Resolution Imaging, and the ONI application note on Quantitative super-resolution reveals nanoscale recruitment of GABAA receptors following Bicuculline treatment.
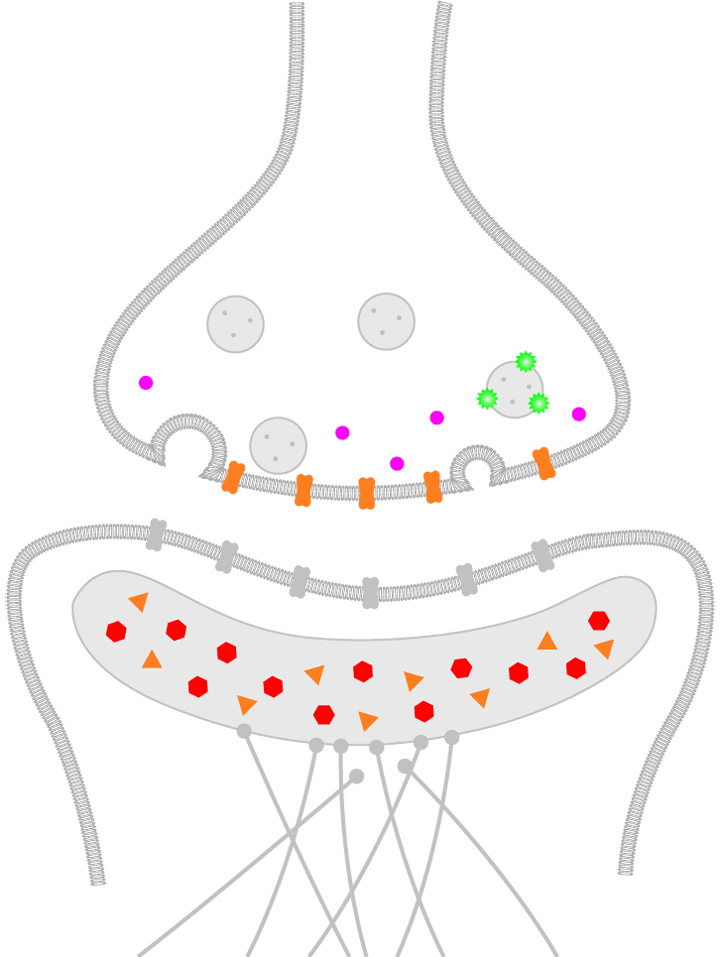
Figure 1. Schematic representation of a human brain synapse. Presynaptic molecules are labeled in purple and green, while the postsynaptic density (PSD) is illustrated with red and orange molecules.
Single-molecule neuroimaging
Because synapses are under 200 nm in size, their intricate molecular details are ‘invisible’ to conventional, diffraction limited light microscopy techniques. Understanding the molecular hallmarks of brain plasticity with single-molecule sensitivity entails characterizing neuronal synapse architecture, and studying the dynamics of synaptic proteins, including molecular interactions and distributions of neurotransmitters, nucleic acids and other biomolecules. Single-molecule imaging is a robust tool for answering the fundamental questions on brain function and dysfunction, particularly when combined with research on the complex interactions within the neuronal ecosystem.
In the presented example, neuronal synapses were visualized in 3-color dSTORM imaging using two presynaptic proteins; Bassoon (synaptic active zone, in magenta) and VGluT1 (synaptic vesicles, in green), and the postsynaptic protein Homer1 (postsynaptic density, in orange). The data revealed the presence and organization of neurotransmitter vesicles in between pre- and postsynaptic protein clusters (Figure 2). To learn more read the ONI application note on revealing synapse architecture at a single-molecule level.
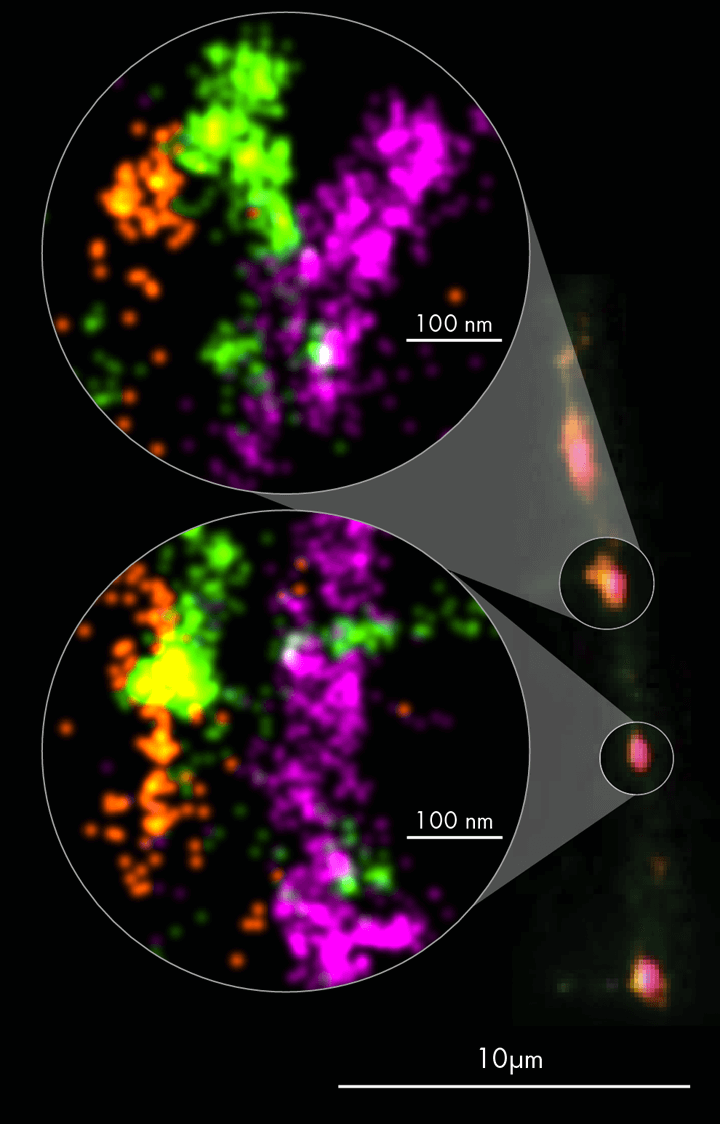
Figure 2. Rat primary cortical neurons were cultured for 14 days, allowing synapse formation on a dish. Neuronal synapses were visualized in 3 color dSTORM imaging using two presynaptic proteins Bassoon (in magenta) and VGluT1 (in green); and the postsynaptic protein Homer1 (in orange). Image credit: ONI Applications Development Team.
Chapter 2
Modeling disease: single-molecule imaging in brain tissues
Brain tissue complexity in neurodegeneration
Our brain is composed of highly complex and densely packed tissues with specialized functions. An adult human brain contains an estimated 86 billion neurons and a similar number of supporting cells, including astrocytes and microglia. On average, each brain cell is connected to around 25,000 other cells, meaning that tissues are integrated by local intertwined cells that project to distant regions. This makes three-dimensional (3D) visualization of brain tissues a challenging job for neuroscientists.
There are several challenges facing research on brain tissues, which commonly include sample sourcing and preparation, specimen labeling, imaging of thick sections, or 3-dimensional data acquisition, often involving time-consuming and laborious protocols. Current efforts are also concentrating on using stem cell models and molecular tools to mimic brain physiology and better explore the origins of neurological disorders.
Current ground-breaking studies are focusing on the discovery and validation of early-disease biomarkers in human fluids, such as cerebrospinal fluid (CSF), which could help diagnose neurodegenerative diseases at early/pre-symptomatic stages, given that high-sensitivity detection of basal levels of biomarkers is achieved (attomolar concentrations in most cases). This would also contribute to the better understanding of disease mechanisms and allow a better patient stratification prior to therapy. Early clinical trials are currently underway to test how small drugs can block or stimulate key molecular interactions involved in neurodegenerative disease progression.
In recent years, extracellular vesicles (EVs) have also been studied as potential carriers of molecules for intercellular communication within tissues and throughout the body, with their unique ability to cross the blood-brain barrier and facilitate communication between peripheral tissues and the central nervous system (CNS). Read our EV guide for more details on single-molecule imaging solutions for studying extracellular vesicles.

Figure 3. Overview scan of a mouse brain tissue section. Image of a 10µm-thick mouse brain tissue. Nuclei were labeled with DAPI and the Nanoimager overview scan feature was used to identify the structure of interest, the hippocampus. Sample prepared by Robert Hinshaw, Ann Romney Center for Neurologic Diseases, Brigham and Women’s Hospital, Boston.
Resolving single synapses in tissue: PALM array tomography
In dense tissue environments, depth often affects light scattering. The brain’s high lipid content (more than half of the solid matter) contributes to the mismatch in refractive index that causes light scattering. Tissue clearing is an approach to counteract this issue, however, it requires specialized training and arduous processing, normally done on >1mm-thick tissue sections.
An approach to tissue imaging that allows molecular and ultrastructure analysis of neuronal circuits in tissues is array tomography. Recent advances in super-resolution microscopy have enabled the development of combined PALM array tomography as a way to image deep tissues with high sensitivity by using physical sectioning, and achieve isotropic high-resolution in all three dimensions.
After identifying a unique structure as the reference point in each of the sections (Figure 4), positions can be imaged in higher detail using PALM to visualize synaptic protein clusters (i.e. PSD95 clusters) and determine their structure and heterogeneity within brain tissues at a single-molecule level (Figure 5). To learn more you can download the ONI application note on resolving single synapses with PALM array tomography.

Figure 4: (LEFT) Overview scan of an ultrathin human brain tissue section labeled with mEOS-tagged PSD95, a postsynaptic protein. This was used to identify a unique structure in each of the sections (A-C), which served as a reference point for PALM Array Tomography imaging. Scale bar: 50 µm. Figure 5: (RIGHT) PALM imaging of human brain tissue labeled with mEOS-tagged PSD95. mEos was photo-converted using UV light. Scale bars: 20nm. Sample prepared by Dr. M. Horrocks, School of Chemistry and Dementia Research Institute, University of Edinburgh.
Labeling single molecules in brain tissues
In vivo labeling of the molecules of interest can also pose a significant bottleneck for researchers, especially when preparing tissue samples for single-molecule imaging. Different labeling approaches can be used for brain tissue in vivo imaging, including photo-activatable fluorescent proteins (FPs) or Halo-tags. Halo-tags combine the advantages of genetically encoded fluorescent proteins with the superior brightness of synthetic dyes and photostability.
The advantage of using an imaging system with simultaneous dual color acquisition is that different labeling methods can be directly compared. In the example, PALM imaging on whole mouse brain tissue slices was used to evaluate the efficiency of in vivo fluorescent labeling of single molecules using Halo tags. The comparison of the localizations between the photo-activated protein PSD95-mEos2 (blue) and the labeled PSD95-Halo tag (magenta) was used to validate the efficiency of Halo-tag labeling in vivo. You can read more about this experiment in the ONI application note on single-molecule in vivo imaging of brain tissues.
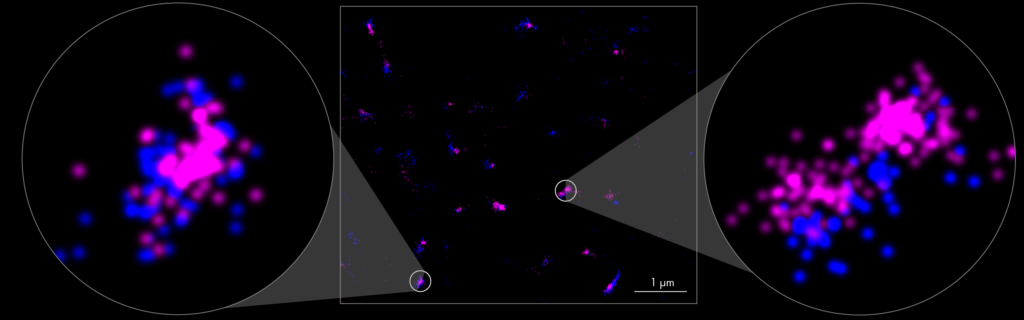
Figure 6. Whole mouse brain tissue labeled with two fluorophores to evaluate in vivo labeling efficiency. PALM image of a 6µm-thick brain tissue section of a PSD95-mEos2/PSD95-Halo mouse (blue and magenta, respectively) labeled with HaloTag-PA Janelia Fluor® 646 dye. Two color PALM imaging was used to assess the labeling efficiency of postsynaptic protein using Halo tag in vivo (zoomed in regions). Sample prepared by Edita Bulovaite (Seth Grant’s group) Centre for Clinical Brain Sciences, University of Edinburgh, UK.
Chapter 3
Neuro-Nanoimaging
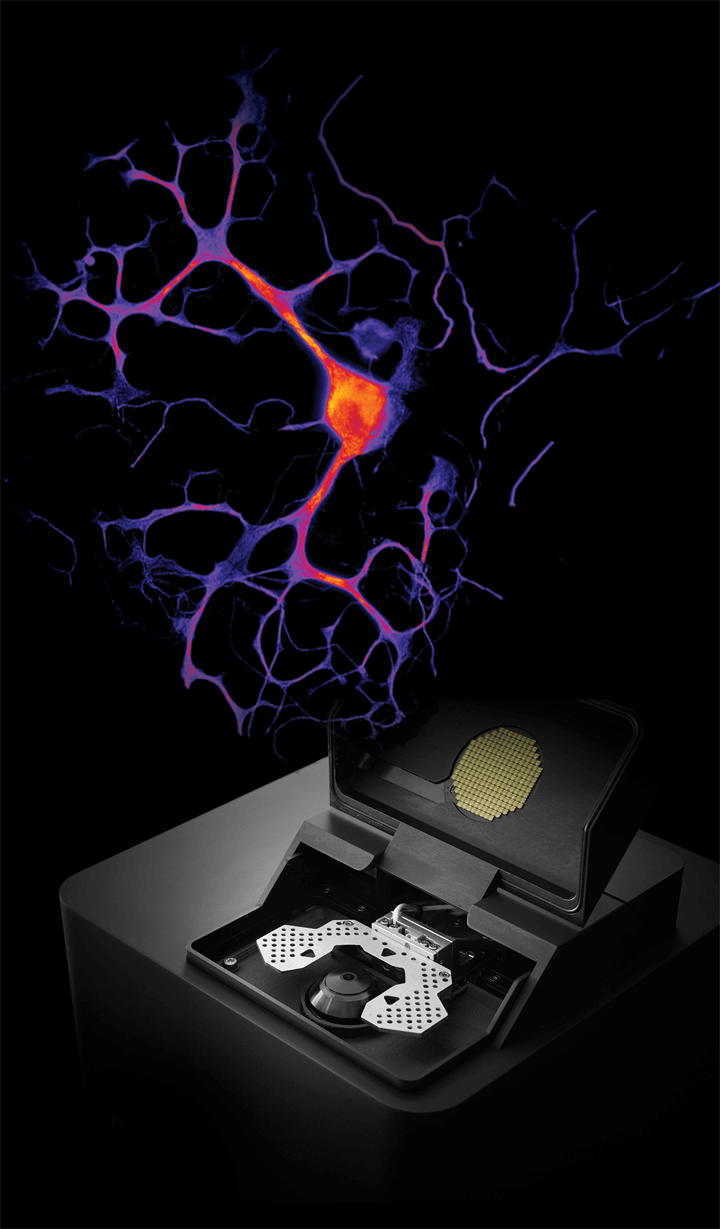
The Nanoimager platform enables single-molecule neuroimaging of cultured neuronal cells and brain tissue sections, with a resolution reaching 20 nm. Combining automation features with super-resolution imaging allows the quick and efficient identification of region of interests to then gather localizations from thousands of synaptic proteins. It also facilitates acquisition of data on single molecule distribution, co-localization, or postsynaptic density profiles.
Supporting both fixed and live sample imaging, researchers can develop their own assays to build a complete spatio-temporal portrait of neuronal pathways. Single-molecule sensitivity means potential for improved, accelerated diagnostics and disease therapies. Understanding how drugs alter neurotransmission mechanisms and kinetics with minimal sample requirements can also translate into robust, fast and efficient drug profiling, in a more cost-efficient manner.
Single-molecule imaging has the potential to help establish and characterize new disease models to enable robust testing of novel therapeutics and facilitate drug development. With its compact design, lack of need for a dark room or additional infrastructure, the Nanoimager is the most accessible super-resolution tool ever designed. To learn more about the microscope and ONI, please visit the Nanoimager page.
Summary
Single-molecule imaging opens new possibilities in neuroimaging field. It gives researchers new tools to:
- Determine synaptic architecture in healthy and disease-affected tissues
- Follow protein dynamics in living brains by imaging different labels simultaneously
- Understand the molecular mechanisms of tissue neurodegeneration
- Identify hallmark disease-linked markers to improve early diagnosis and provide new drug targets
- Speed up drug development and characterization of neurodegenerative, psychiatric or neurodevelopmental disorders