Introducing: ONI’s Next-Generation End-to-End Solution for EV Research Learn More
DNA Conformational Changes
Challenge
Understanding protein interactions
The interactions of the protein factors at the nanoscale plays a very important role in deciding the fate of cellular processes of an organism. Conventionally, images of cells are used to understand the relationship between molecules through their spatial organization. However, often the most important aspect of an interaction is the rate of binding, the dissociation constant, conformational change and other dynamics and dwell times. This information is especially useful for understanding the effect of inhibitors and small molecules or solution conditions on the behavior of a particular interaction, such as the activity of an enzyme induced by substrate binding.
Single-molecule Förster Resonance Energy Transfer (smFRET) is a technique based on the non-radiative energy transfer between two fluorescently labeled molecules, a donor and an acceptor. When the distance between the donor fluorophore and the acceptor fluorophore is lower than 10 nm, upon excitation of the donor fluorophore by the laser, the energy from the donor fluorophore is transferred to the acceptor fluorophore. The efficiency of energy transfer is correlated to the distance between the donor and acceptor fluorophores. Thus smFRET acts as a formidable real time nanoruler within the 2-10 nm range, to understand dynamic biological processes such as transcription of a gene.
In spite of its robustness as a tool to probe and manipulate biological nano machines, smFRET has largely remained a technique solely used by researchers with biophysical expertise.
Solution with the Nanoimager
Researching biological systems with smFRET
The low adoption rate of smFRET by biological researchers can largely be blamed on the lack of a user-friendly commercial instrument, together with the steep learning curve, expensive equipment and required expertise in data analysis.
The Nanoimager, with a simple design and a user friendly interface, makes it easier for anyone to perform smFRET experiments. smFRET can characterize single-molecule interactions and quantitate them for various types of sample: molecules bound to a surface, in solution, on an artificial membrane or on a live cell membrane close to a glass coverslip. All experiments can be done just by click of a button without any prior experience with NimOS smFRET data acquisition or analysis.
Case Study
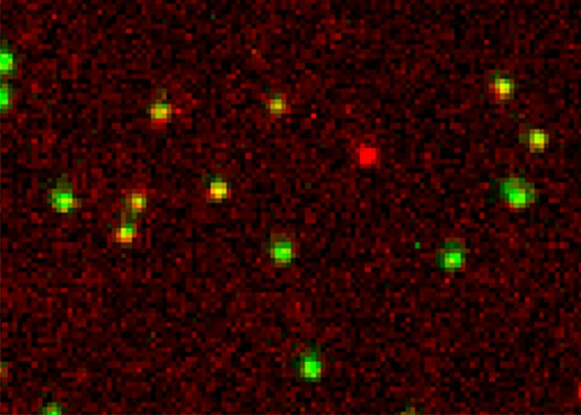
Using smFRET to study Holliday junctions
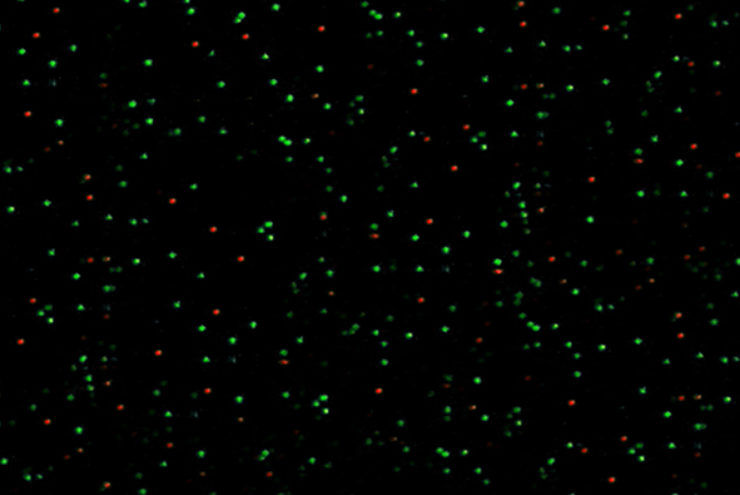
A Holliday junction is a very dynamic DNA structure formed during DNA recombination inside the cell. Here we reconstituted this dynamic structure using synthetic DNA to understand the dynamics using single-molecule FRET. The structure was immobilized on to a passivated glass surface by biotin. In green is the intensity of donor fluorophore and in red is the intensity of the acceptor fluorophore. The different conformation of the Holliday junction can be measured in real time with this smFRET experiment.
Learn more about single-molecule FRET or read about other super-resolution microscopy techniques supported by the Nanoimager below.